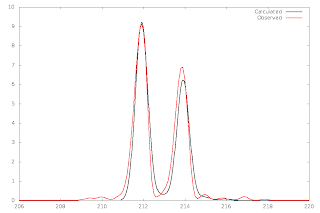
In part 2, I showed you how to generate theoretical isotopic envelopes, and how to compare them with observed ones: http://verahill.blogspot.com.au/2013/07/480-ms-data-part-ii-plotting-and.html
In part 3, I showed you how to make contour plots of '3D' data -- in that particular case the extra dimension was cone voltage, but it could just as easily have been time: http://verahill.blogspot.com.au/2013/07/ms-data-part-iii-generating-matrix-by.html
In part 4, we'll use the same data as in part 3, but we'll make a stacked plot of it. This is a short post.
There is at least one other way of making a stacked plot in gnuplot, but it doesn't yield what I'd consider as being publication quality plots. It's also fairly restrictive in the type of data can be plotted. The method shown here is general and applicable to all types of data. You can e.g. use it for time-dependent NMR data...
Here's an example of a gnuplot script:
### Preamble start set term png size 1000,500 set output 'stack.png' set border 1 set xtics nomirror set ytics nomirror unset ytics set xrange [100:3000] set yrange [0:100] set multiplot set size 0.85,0.45 unset key ### Preamble over ### The stacked plot set origin 0,0 set label "m/z" at 1500, -18 plot '0.dat' with lines lt -1 # we want the x axis to show ONLY for the first spectrum, # so we turn it off for the remaining spectra. # The same goes for our label (xlabel can be tricky # to position correctly) unset label set border 0 unset xtics unset ytics # Here we set the spacing (fx) the initial position of the second spectrum (f), # the initial horizontal offset of the second spectrum relative to the first one (g), # and the horizontal offset for all subsequent spectra (gy) f=0.0025 fx=0.00 g=0.01 gy=0.02 # then we plot all the other spectra set origin fx+0*f,gy+0*g plot '10.dat' with lines lt -1 set origin fx+1*f,gy+1*g plot '20.dat' with lines lt -1 set origin fx+2*f,gy+2*g plot '30.dat' with lines lt -1 set origin fx+3*f,gy+3*g plot '40.dat' with lines lt -1 set origin fx+4*f,gy+4*g plot '50.dat' with lines lt -1 set origin fx+5*f,gy+5*g plot '60.dat' with lines lt -1 set origin fx+6*f,gy+6*g plot '70.dat' with lines lt -1 set origin fx+7*f,gy+7*g plot '80.dat' with lines lt -1 set origin fx+8*f,gy+8*g plot '90.dat' with lines lt -1 set origin fx+9*f,gy+9*g plot '100.dat' with lines lt -1 set origin fx+10*f,gy+10*g plot '110.dat' with lines lt -1 set origin fx+11*f,gy+11*g plot '120.dat' with lines lt -1 set origin fx+12*f,gy+12*g plot '130.dat' with lines lt -1 set origin fx+13*f,gy+13*g plot '140.dat' with lines lt -1 set origin fx+14*f,gy+14*g plot '150.dat' with lines lt -1 set origin fx+15*f,gy+15*g plot '160.dat' with lines lt -1 set origin fx+16*f,gy+16*g plot '170.dat' with lines lt -1 set origin fx+17*f,gy+17*g plot '180.dat' with lines lt -1 set origin fx+18*f,gy+18*g plot '190.dat' with lines lt -1 set origin fx+19*f,gy+19*g plot '200.dat' with lines lt -1 set origin fx+20*f,gy+20*g plot '210.dat' with lines lt -1 set origin fx+21*f,gy+21*g plot '220.dat' with lines lt -1 set origin fx+22*f,gy+22*g plot '230.dat' with lines lt -1 set origin fx+23*f,gy+23*g plot '240.dat' with lines lt -1 set origin fx+24*f,gy+24*g plot '250.dat' with lines lt -1 set origin fx+25*f,gy+25*g plot '260.dat' with lines lt -1 set origin fx+26*f,gy+26*g plot '270.dat' with lines lt -1 set origin fx+27*f,gy+27*g plot '280.dat' with lines lt -1 set origin fx+28*f,gy+28*g plot '290.dat' with lines lt -1 set origin fx+29*f,gy+29*g plot '300.dat' with lines lt -1
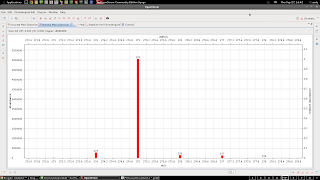
and here's the plot:
If you want to make it really fancy, try this:
### Preamble start set term png size 1000,800 set output 'stack.png' set border 1 set xtics nomirror set ytics nomirror unset ytics set xrange [100:3000] set yrange [0:100] set multiplot set size 0.85,0.25 unset key ### Preamble over ### The stacked plot set origin 0,0 set label "m/z" at 1500, -18 plot '0.dat' with lines lt -1 # we want the x axis to show ONLY for the first spectrum, # so we turn it off for the remaining spectra. # The same goes for our label (xlabel can be tricky # to position correctly) unset label set border 0 unset xtics unset ytics # Here we set the spacing (fx) the initial position of the second spectrum (f), # the initial horizontal offset of the second spectrum relative to the first one (g), # and the horizontal offset for all subsequent spectra (gy) f=0.0025 fx=0.00 g=0.01 gy=0.02 # then we plot all the other spectra set origin fx+0*f,gy+0*g plot '10.dat' with lines lt -1 set origin fx+1*f,gy+1*g plot '20.dat' with lines lt -1 set origin fx+2*f,gy+2*g plot '30.dat' with lines lt -1 set origin fx+3*f,gy+3*g plot '40.dat' with lines lt -1 set origin fx+4*f,gy+4*g plot '50.dat' with lines lt -1 set origin fx+5*f,gy+5*g plot '60.dat' with lines lt -1 set origin fx+6*f,gy+6*g plot '70.dat' with lines lt -1 set origin fx+7*f,gy+7*g plot '80.dat' with lines lt -1 set origin fx+8*f,gy+8*g plot '90.dat' with lines lt -1 set origin fx+9*f,gy+9*g plot '100.dat' with lines lt -1 set origin fx+10*f,gy+10*g plot '110.dat' with lines lt -1 set origin fx+11*f,gy+11*g plot '120.dat' with lines lt -1 set origin fx+12*f,gy+12*g plot '130.dat' with lines lt -1 set origin fx+13*f,gy+13*g plot '140.dat' with lines lt -1 set origin fx+14*f,gy+14*g plot '150.dat' with lines lt -1 set origin fx+15*f,gy+15*g plot '160.dat' with lines lt -1 set origin fx+16*f,gy+16*g plot '170.dat' with lines lt -1 set origin fx+17*f,gy+17*g plot '180.dat' with lines lt -1 set origin fx+18*f,gy+18*g plot '190.dat' with lines lt -1 set origin fx+19*f,gy+19*g plot '200.dat' with lines lt -1 set origin fx+20*f,gy+20*g plot '210.dat' with lines lt -1 set origin fx+21*f,gy+21*g plot '220.dat' with lines lt -1 set origin fx+22*f,gy+22*g plot '230.dat' with lines lt -1 set origin fx+23*f,gy+23*g plot '240.dat' with lines lt -1 set origin fx+24*f,gy+24*g plot '250.dat' with lines lt -1 set origin fx+25*f,gy+25*g plot '260.dat' with lines lt -1 set origin fx+26*f,gy+26*g plot '270.dat' with lines lt -1 set origin fx+27*f,gy+27*g plot '280.dat' with lines lt -1 set origin fx+28*f,gy+28*g plot '290.dat' with lines lt -1 set origin fx+29*f,gy+29*g plot '300.dat' with lines lt -1 ### second plot set size 0.75,0.4 set origin 0.1,0.6 set xrange [800:1800] set border 3 set xtics nomirror set ytics nomirror set xlabel 'm/z' set ylabel 'Relative abundance (%)' set title '20 V' plot '20.dat' with lines lt -1 unset multiplot
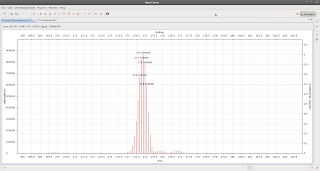
which looks like this: